BLAST, which stands for Basic Local Alignment Search Tool, is a commonly used software tool and algorithm for comparing basic biological sequence information, such as the amino-acid sequences of proteins or the nucleotide (ATGC) sequences of DNA or RNA. It’s a fundamental tool in bioinformatics and molecular biology research, aiding in tasks such as identifying homologous sequences, determining evolutionary relationships, and annotating genomic sequences.
BLAST compares a query sequence (the sequence being searched) against a database of sequences. The algorithm looks for regions of similarity, called alignments, between the target sequence and sequences in the database. It then ranks the database sequences based on the degree of similarity to the query sequence.
Key Features of Basic Local Alignment Search Tool
Critical features of BLAST include:
- Speed: BLAST is designed to rapidly search large sequence databases, efficiently analyzing genomic and proteomic data.
- Sensitivity: BLAST can detect remote homologs with significant sequence similarity, even when they share only a tiny proportion of identical residues.
- Scoring System: BLAST uses a scoring system to evaluate sequence alignments, considering factors such as matches, mismatches, gap openings, and gap extensions. This scoring system helps determine the significance of sequence similarities.
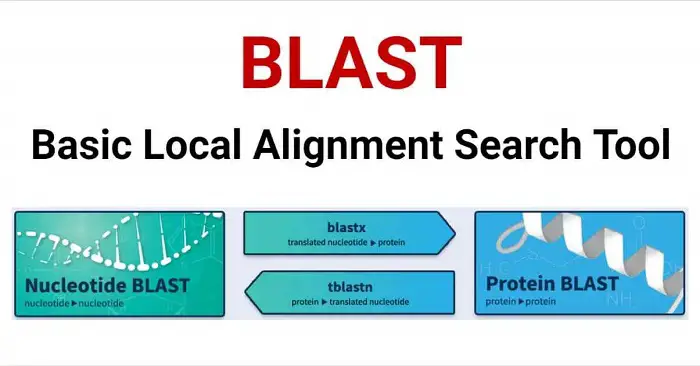
- Various Algorithms: There are different variants of BLAST tailored for specific types of sequences and databases, such as BLASTP (for protein sequences), BLASTN (for nucleotide sequences), BLASTX (translates nucleotide sequences in all six frames and compares them to protein sequences), and others.
BLAST is widely used in bioinformatics for a variety of purposes, including:
- Identifying genes and proteins with similar functions across different species.
- Annotating newly sequenced genomes by comparing them to known sequences in databases.
- Investigating evolutionary relationships by comparing sequences from different organisms.
- Predicting the function of proteins based on sequence similarity to known proteins.
- Identifying potential drug targets by comparing the protein sequences of pathogens to those of the host organism.
Overall, BLAST is a powerful and versatile tool that has revolutionized how researchers analyze biological sequence data.
Types of Basic Local Alignment Search Tool (BLAST)
There are several variants of BLAST, each optimized for specific types of biological sequences and databases. The most commonly used types of BLAST include:
- BLASTP: This variant compares a protein query sequence against a protein sequence database. BLASTP is commonly used for protein homology searches and for identifying similar proteins with known functions.
- BLASTN: BLASTN compares a nucleotide query sequence against a nucleotide sequence database. It is commonly used for nucleotide sequence homology searches, such as identifying similar DNA or RNA sequences.
- BLASTX: BLASTX compares a nucleotide query sequence translated in all six reading frames against a protein sequence database. This allows for the identification of potential protein-coding regions in nucleotide sequences.
- tBLASTN: This variant compares a protein query (amino-acid) sequence against a nucleotide (ATGC) sequence database translated in all six reading frames. It helps identify potential protein-coding regions in DNA sequences.
- tBLASTX: tBLASTX compares the six-frame translations of a nucleotide (ATGC) query sequence against the six-frame translations of a nucleotide sequence database. This allows for the detection of distant relationships between nucleotide sequences.
These variants of BLAST cover a wide range of sequence comparison scenarios and are essential tools in bioinformatics and molecular biology research. Researchers can choose the appropriate type of BLAST-based on the nature of their query sequence and the type of database they wish to search against.